
This way, the solution becomes quadratic O(n^2) time where n is length of the string. So, cache those results and reuse them for queries generated by other 2 character matches. It is also known as a palindrome or an inverted-reverse sequence.

This definition of palindrome thus depends on complementary strands being palindromic of. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. Hence, the answer is 2.Ĭomputing how many palindromes of length 3 are possible for every substring can be time consuming if we don't cache the results when we do it the first time. A palindromic sequence is a sequence made up of nucleic acids within double helix of DNA and/or RNA that is the same when read from 5 to 3 on one strand and 5 to 3 on the other, complementary, strand. A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. Stop transcription and initiate translation. Hence, we can make palindromic sequence of length 5 as abcba or acbca. Importance of Palindromic sequences: Gene expression and regulation Gene replication Homologous recombination The recognition site for restriction enzymes Binding sites for many enzymes and transcriptional factors. Due to the palindromic sequences often found on ChrY, Y-STR loci can sometimes exhibit as. In the above example, you can find 2 palindromes of length 3 which is bcb and cbc. A palindromic sequence of 146 nt is at the 3-end of the genome and a different palindromic sequence of. Whenever 2 characters match, we only have to find how many palindromes of length 3 are possible in between these 2 characters. Palindromic sequence-targeted (PST) PCR: a rapid and efficient method for high-throughput gene characterization and genome walking Genome walking (GW) refers to the capture and sequencing of unknown regions in a long DNA molecule that are adjacent to a region with a known sequence.When two carbohydrates are epimers: A) one is a pyranose, the other a furanose. Which of the following monosaccharides is not an aldose? A) erythrose B) fructose C) glucose D) glyceraldehyde E) ribose 9. If last and first characters of X are same, then L(0, n-1) L(1, n-2) + 2. E) the template DNA strand is radioactive. Let X0.n-1 be the input sequence of length n and L(0, n-1) be the length of the longest palindromic subsequence of X0.n-1. For example, restriction enzymes often recognize palindromic sequences of DNA. They occur in genomes of all organisms and have various functions.

The new method uses less memory and thereby it increases the overall speed and efficiency. Palindromes in DNA consist of nucleotides sequences that read the same from the 5'-end to the 3'-end, and its double helix is related by twofold axis. Thus, a new method has been developed using dynamic programming to fetch the palindromic nucleic acid sequences. For example, the sequence GGATCC on one strand of DNA is considered a palindrome because the sequence on its complementary strand is CCTAGG. D) the role of the dideoxy CTP is to occasionally terminate enzymatic synthesis of DNA where Gs occur in the template strands. A typical nucleotide palindromic sequence would be TATA (5' to 3') and its complimentary sequence from 3' to 5' would be ATAT. A palindromic sequence of nucleotides (which are labeled A, T, C, or G) occurs when complementary strands of DNA read the same in both directions, either from the 5-prime end or the 3-prime end. B) specific enzymes are used to cut the newly synthesized DNA into small pieces, which are then separated by decrophesis C) the dideoxynucleotides must be present at high levels to obtain long stretches of DNA sequence. In DNA sequencing by the Sanger (dideaxyl method: A) radioactive dideoxy ATP is included in each of four reaction mixtures before enzymatic synthesis of complementary strands.
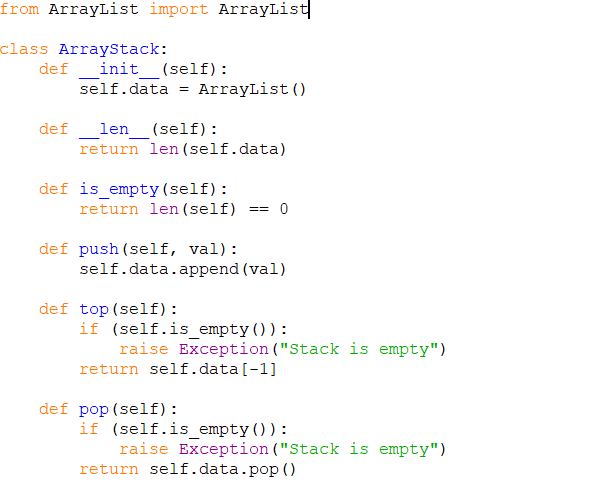
Calculate the reverse complement of the whole forward strand to finish. Reverse the order of the reverse complement ( CTA) and add it to the end of the forward strand - TAGCTA. Calculate the reverse complement of the sequence - ATC. Which of the following deoxyoligonucleotides will hybridize with a DNA containing the sequence (5)AGACTGGTC(3)? A) (5)CTCATTGAG(3) B) (5)GACCAGTCT(3) C) (5)GAGTCAAC (3) D) (5)TCTGACCAG(3) E) (5)TCTGGATCT(3) 7. Starting with the original sequence - TAG. Palindromic sequences are the usual recognition sites for restriction enzymes and frequently occur as essential elements in regulatory regions. In this problem, one sequence of characters is given, we have to find the longest length of a palindromic subsequence. Certain DNA sequences have a twofold inverted symmetry and are called self-complementary or palindromic sequences. Which of the following is a palindromic sequence? A) AGGTCC TCCAGG B) CCTTCC GCAAGG C) GAATCC CTTAGG D) GGATCC CCTAGG E) GTATCC CATAGG 6. Longest Palindromic Subsequence is the subsequence of a given sequence, and the subsequence is a palindrome.


 0 kommentar(er)
0 kommentar(er)
